 Discovery and characterization of somatic genomic structural variation in human tissues
Discovery and characterization of somatic genomic structural variation in human tissues
The primary purpose of sequencing genomes is to identify the underlying genetic variation between individuals and to explore what role those changes have on human phenotypes. Our research laboratory develops and implements methods to precisely identify and resolve different types of genomic variation both between and within individuals. Our current efforts are focused on developing methods for discovering and characterizing somatic genomic variation in individual cells and tissues. Our goal is to integrate this information with other forms of biologically and medically relevant data to improve our overall understanding of human health and disease.
Over the past decade, our research has been conducted independently, collaboratively through partnerships with other research groups, and through membership in numerous large consortia including the 1000 Genomes Project, Human Genome Structural Variation Consortium, Brain Somatic Mosaicism Network, Impact of Genomic Variation on Function Consortium, and recently the Somatic Mosaicism across Human Tissues Network.
Areas of Investigation
1. Discovery and Analysis of Structural Genomic Variation in Human Populations: Our interest in structural variation began with early work constructing some of the earliest maps of insertion/deletion variation as well as the first large-scale sequence-based assessment of copy number variation across multiple human populations. Our current research has continued in this area, where we have published several methods for identifying different types of structural variation from whole genome sequencing including copy number variation, complex rearrangements, nuclear mitochondrial insertions, and mobile element insertions. We continue to progress in these areas, with ongoing projects studying allelic heterogeneity in polymorphic mobile element insertions and modeling the potential impact of structural variation on gene expression and chromatin structure. We are also investigating short tandem repeat expansions in large cohorts of individuals with neurodevelopment and neuropsychiatric disorders and have been developing cyberinfrastructure for low-cost/high-throughput analysis using Amazon Web Services (AWS) and other cloud-based systems.
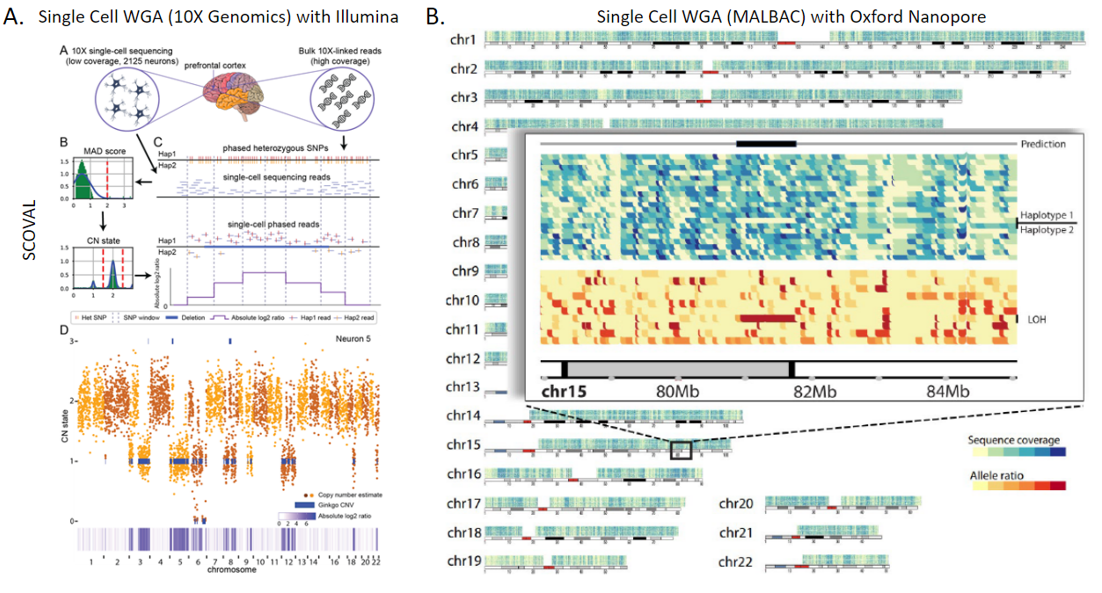
2. Technologies to Identify Somatic Structural Variation in Human Tissues: While there are many studies on somatic structural variation in cancers, there are far fewer that explore their prevalence and impact within non-cancerous human tissues. This has been in part due to limitations of technology, limiting investigations to either higher frequency mosaicism or very large events detectable by microscopy or fluorescence in situ hybridization (FISH) based assays. Our group became involved in this area of research through our participation in the Brian Somatic Mosaicism Network (BSMN), a consortium formed to explore different types of genetic variation within human brain tissue. We helped lead initiatives to explore best practices in somatic SNV calling as well as somatic copy number variation from whole genome amplified single cell. In parallel to our BSMN-related research, we have also been exploring the somatic prevalence of nuclear mitochondrial insertions in the aging human brain. This work in aggregate, coupled with other ongoing research into somatic mobile element insertions, led to a recent award as part of the new Somatic Mosaicism across Human Tissues (SMaHT) consortium to continue to explore this type of variation in other human tissues by developing new tools and technologies.
3. Characterization and Impact of Human Papilloma Virus (HPV) in Head and Neck Cancers: Our interest in structural variation has further led to an ongoing collaboration on HPV integrations in head and neck squamous cell carcinomas (HNSCC). When HPV integrates into a human genome, it is often associated with structural rearrangements around the insertion site which can result in a focal amplification of the region, resulting in sometimes over 100 copies of the HPV element and surround area. This is likely due to a rolling circle amplification of the HPV genome, and while such amplifications are not present in every HPV-associated HNSCC, we hypothesize that different integration structures are associated with distinct mechanisms for carcinogenesis and progression. Our group developed an approach, SearcHPV, to identify HPV integrations and resulting rearranged genomic structures from targeted capture approaches and have contributed in its application to explore HPV characteristics as a biomarker and its role in mucoepidermoid carcinoma.
